Introduction
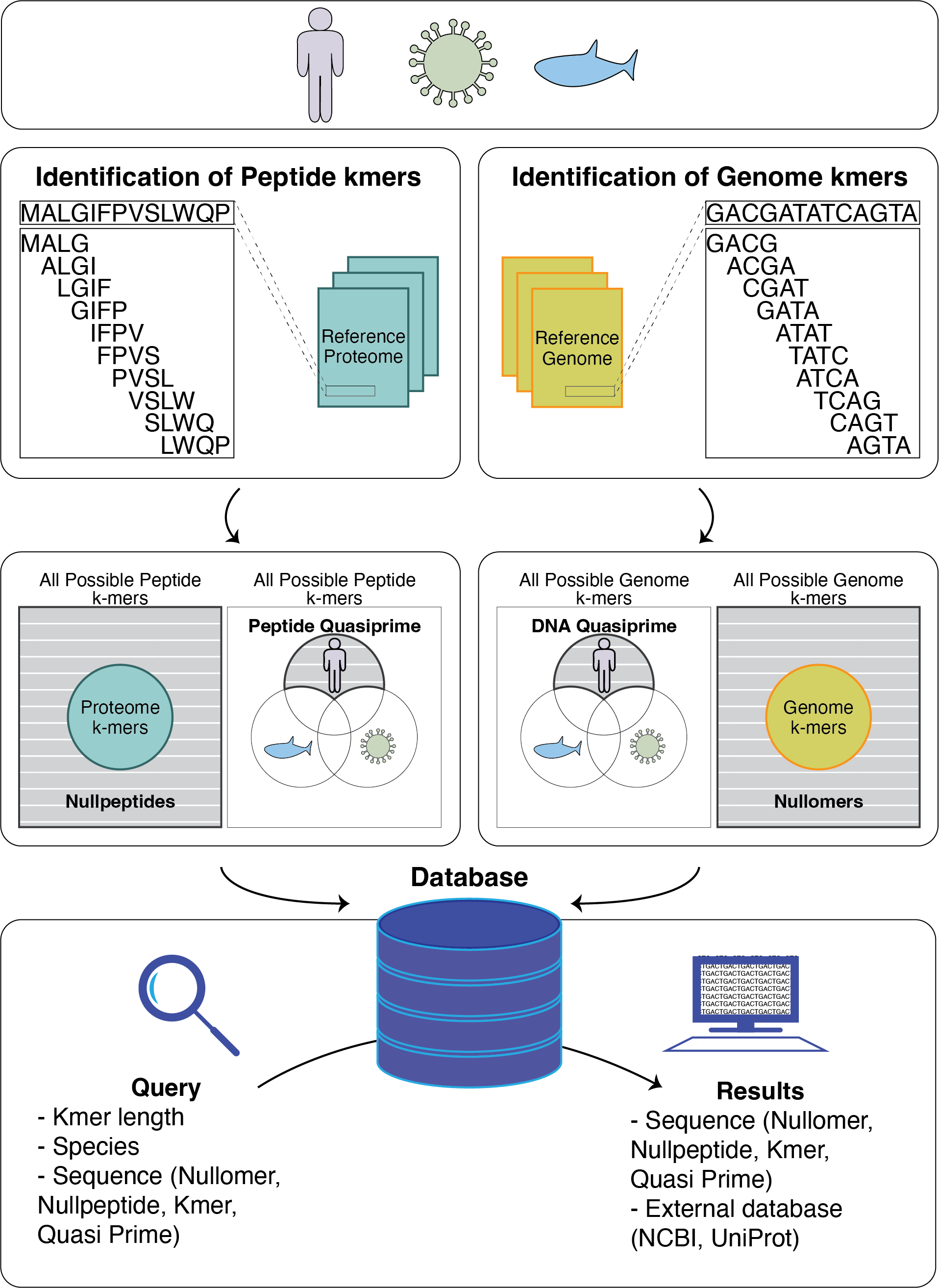
kmerDB is a comprehensive portal for DNA and peptide sequences. The database contains the kmers found in X reference genomes and 22,386 reference proteomes. It also encompasses the kmers absent from reference genomes and proteomes, known as nullomers and nullpeptides, respectively. The database is also subdivided by taxonomy in eukaryotes, archaea, bacteria and viruses. Quasi-primes are sequences that are unique in a species. kmerDB contains the set of quasi-primes of each reference genome and proteome.
The database can be searched based on species name, ID or based on the sequence the user is interested in. Advanced searches can use the number of kmers or nullomers in a species for searching. kmerDB also provides integration with NCBI and UniProt databases.
Definitions
-
Kmers: Contiguous subsequences of length k that are derived from longer sequences.
-
Nullomers: Kmer sequences that do not exist within a certain genome.
-
Primes: kmers that do not exist in any genome or proteome.
-
Nullpeptides: Kmer sequences that do not exist within a certain proteome.
-
Quasi-Primes: Kmer sequences found only in one species and are absent in every other genome or proteome
References
-
Mouratidis, Ioannis, Candace SY Chan, Nikol Chantzi, Georgios Christos Tsiatsianis, Martin Hemberg, Nadav Ahituv, and Ilias Georgakopoulos-Soares. "Quasi-prime peptides: identification of the shortest peptide sequences unique to a species." NAR Genomics and Bioinformatics 5, no. 2 (2023): lqad039.
-
Georgakopoulos-Soares, Ilias, Ofer Yizhar-Barnea, Ioannis Mouratidis, Martin Hemberg, and Nadav Ahituv. "Absent from DNA and protein: genomic characterization of nullomers and nullpeptides across functional categories and evolution." Genome biology 22 (2021): 1-24.
To cite kmerDB:
Mouratidis, I., Baltoumas, F.A., Chantzi, N., Chan, C.S.Y., Montgomery, A., Konnaris, M., Georgakopoulos, G.C., Chartoumpekis, D., Kovac, D., Pavlopoulos, G.A. & Georgakopoulos-Soares, I. (2023) “kmerDB: a database encompassing the set of genomic and proteomic sequence information of each species.”
Contact Information
Contact us for feature requests, bugs or collaborations: izg5139@psu.edu pavlopoulos@fleming.gr